Phylogeny and Taxonomy
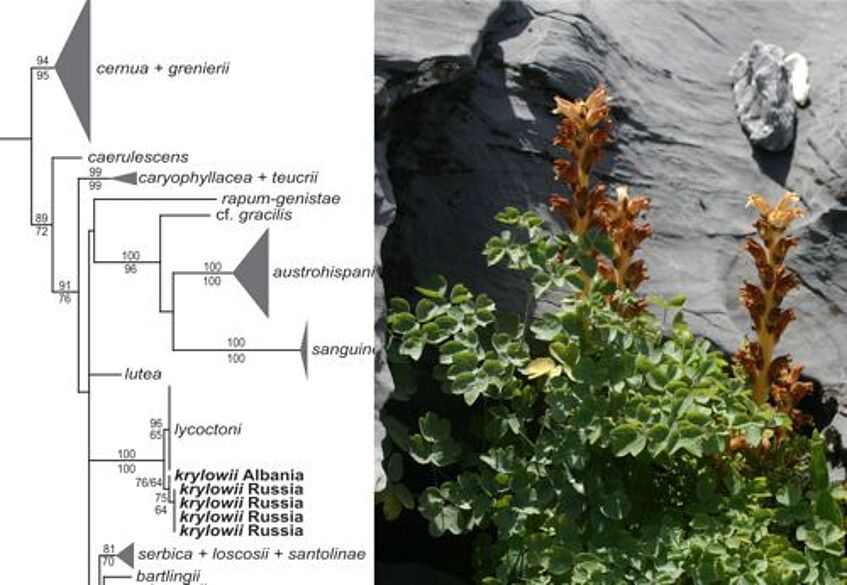
Phylogenetic position of Orobanche krylowii from Albania (photo: B Frajman)
Phylogeny and Taxonomy
In the last years enormous progress has been made with respect to our understanding of circumscription of major clades and of genera within the broomrape family. However, due to incomplete sampling (e.g., about one third of Orobanchaceae genera has not been investigated yet with molecular phylogenetic tools) and varied issues of the used markers (e.g., insufficient resolution, phylogenetic incongruence), some of which are parasite-specific (e.g., absence of some genes in the reduced plastid genomes of non-photosynthetic lineages), numerous open questions remain. Applying commonly used as well as newly developed markers, we are addressing relationships among major clades in Orobanchaceae. Furthermore, we have developed a bioinformatic pipeline for identification of markers to be used in a target-capture phylogenomic approach.
On the genus level, our main interest is on Orobanche and related genera. Using molecular-phylogenetic approaches, occasionally complemented by karyological, cytological and morphological data, we are working on taxonomic questions both at the genus level (e.g., resurrection of a narrow generic concept in Orobanche sensu lato, phylogenetic position of thus far unstudied genera) and the species level (e.g., phylogenetic positions of recently described species).
Selected Publications
- Li X, Feng T, Randle C, Schneeweiss GM (2019) Phylogenetic relationships in Orobanchaceae inferred from low-copy nuclear genes: consolidation of major clades and identification of a novel position of the non-photosynthetic Orobanche clade sister to all other parasitic Orobanchaceae. Frontiers in Plant Science 10: 902
- Li X, Jang TS, Temsch EM, Kato H, Takayama K, Schneeweiss GM (in press) Molecular and karyological data confirm that the enigmatic genus Platypholis from Bonin-Islands (SE Japan) is phylogenetically nested within Orobanche (Orobanchaceae). Journal of Plant Research 130: 273–280
- Schneider AC, Colwell AEL, Schneeweiss GM, Baldwin BG (2016) Extensive cryptic host-specific diversity among western hemisphere broomrapes (Orobanche s.l., Orobanchaceae). Annals of Botany 118: 1101–1111
- Frajman B, Carlón L, Kosachev P, Sánchez Pedraja Ó, Schneeweiss GM, Schönswetter P (2013) Phylogenetic position and taxonomy of the enigmatic Orobanche krylowii (Orobanchaceae), a predominantly Asian species newly found in Albania (SE Europe). Phytotaxa 137 (1): 1–14
- Schneeweiss GM (2013) Phylogenetic relationships and evolutionary trends in Orobanchaceae. Pp 243–265 in: Joel DM, Gressel J, Musselman LJ (eds.): Parasitic Orobanchaceae. Wien, Heidelberg, New York: Springer.
- Schneeweiss GM, Colwell AE, Park J-M, Jang CG, Stuessy TF (2004) Phylogeny of holoparasitic Orobanche (Orobanchaceae) inferred from nuclear ITS-sequences. Molecular Phylogenetics and Evolution 30: 465–478
Genome Evolution
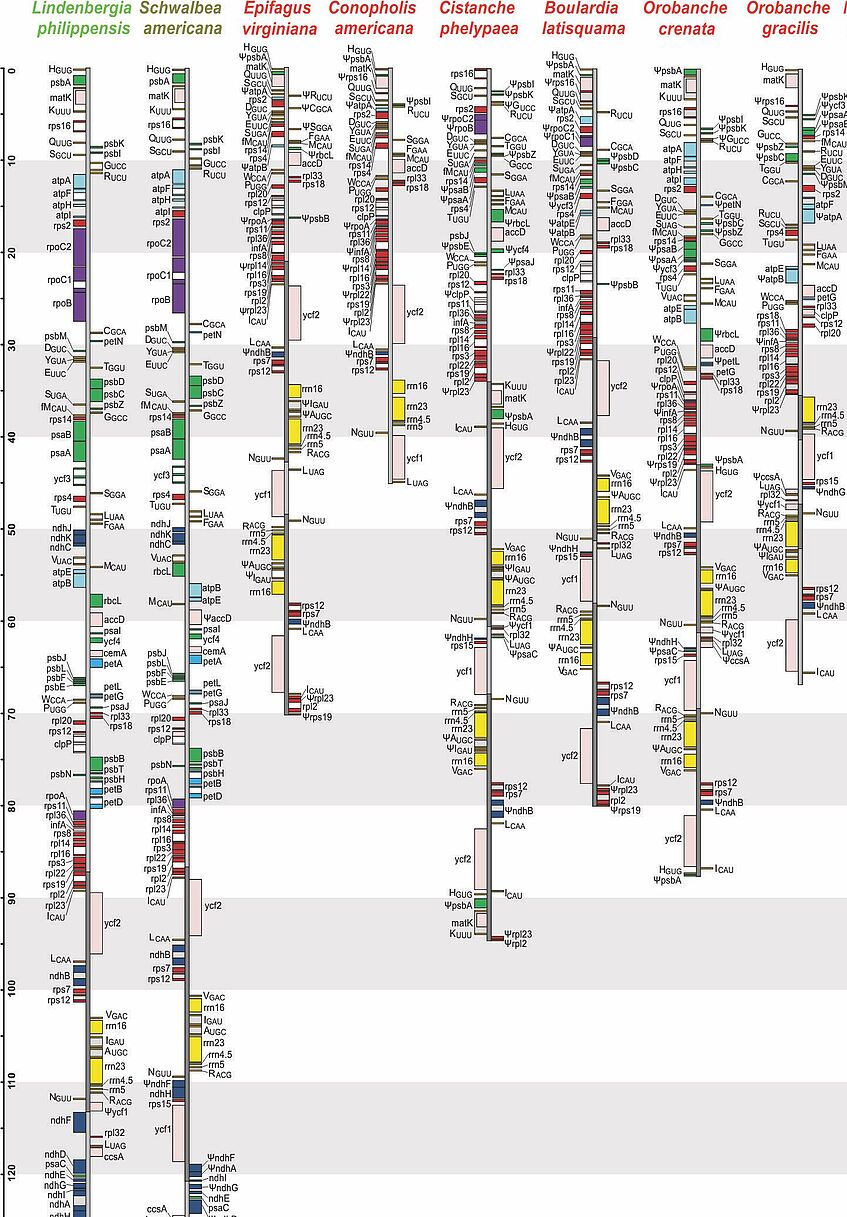
Plastid genomes of Orobanchaceae
Genome Evolution
In the majority of angiosperms, plastid genomes are conservative with respect to structure and gene content. A few groups, however, have undergone major changes, the most prominent being non-photosynthetic parasitic plants. Having abandoned photosynthesis altogether, the genes for photosynthesis have lost their function causing their pseudogenization and eventual loss, but such changes also affect other plastid regions. We have shown that already the establishment of obligate parasitism triggers the relaxation of selective constraints. The loss of photosynthesis alters the chromosomal architecture in that recombinogenic factors accumulate, fostering large-scale chromosomal rearrangements as functional reduction proceeds. The retention of plastid DNA fragments is strongly influenced by both their proximity to genes under selection and the co-occurrence with those in operons, indicating complex constraints beyond gene function that determine the evolutionary survival time of plastid regions in nonphotosynthetic plants. On the level of selection, we have found that relaxed purifying selection in plastid genes is linked already to the origin of obligate parasitism, long before the loss of photosynthesis. Bursts of gene losses coincide with periods of relaxed selection, which are followed by phases of intensified selection and rate deceleration in the retained functional complexes, resulting in “roller-coaster rate variation” along the transition to full parasitism.
Parasitic plants are a rewarding object for studying nuclear genome evolution. Compared to photosynthetic non-parasites and parasites, non-photosynthetic parasitic species contain higher proportions of repetitive DNA sequences, possibly reflecting relaxed selection on genome size in parasitic organisms. Additional support for this "reduced genome economy" hypothesis comes from chromosome number data, because non-photosynthetic parasites tend to have higher chromosome numbers and ploidy levels than their relatives. Genome dynamics may be mostly lineage-specific, as was found for the two broomrape genera Orobanche and Phelipanche, although associations with life traits, such as host range or life history, may exist. Parasitic plants have been identified as important vectors for horizontal gene transfer, even between parasites, adding an additional layer of genomic complexity.
Selected Publications
- Wicke S, Müller KF, dePamphilis CW, Quandt D, Bellot S, Schneeweiss GM (2016) Mechanistic model of evolutionary rate variation en route to a nonphotosynthetic lifestyle in plants. Proceedings of the National Academy of Sciences USA 113: 9045–9050
- Wicke S, Müller KF, dePamphilis CW, Quandt D, Wickett NJ, Zhang Y, Renner SS, Schneeweiss GM (2013) Mechanisms of functional and physical genome reduction in photosynthetic and nonphotosynthetic parasitic plants of the broomrape family (Orobanchaceae). The Plant Cell 25: 3711–3725
- Piednoël M, Aberer AJ, Schneeweiss GM, Macas J, Novak P, Gundlach H, Temsch EM, Renner SS (2012) Next-generation sequencing reveals the impact of repetitive DNA across phylogenetically closely related genomes of Orobanchaceae. Molecular Biology and Evolution 29: 3601–3611
- Park J-M, Manen J-F, Schneeweiss GM (2007) Horizontal gene transfer of a plastid gene in the non-photosynthetic flowering plants Orobanche and Phelipanche (Orobanchaceae). Molecular Phylogenetics and Evolution 43: 974–985
- Schneeweiss GM, Palomeque T, Colwell AE, Weiss-Schneeweis H (2004) Chromosome numbers and karyotype evolution in holoparasitic Orobanche (Orobanchaceae) and related genera. American Journal of Botany 91: 439–448
